Bioactive secondary metabolites (natural products) are produced by every domain of life, ranging from bacteria, to plants, and to animals. Importantly, they have been critical to the development of new drugs, with nearly 50% of FDA approved drugs having their roots in natural product chemistry. Moreover, detailed study of the biosynthesis of many classes of natural products has revealed exciting enzymology not observed in primary metabolism.
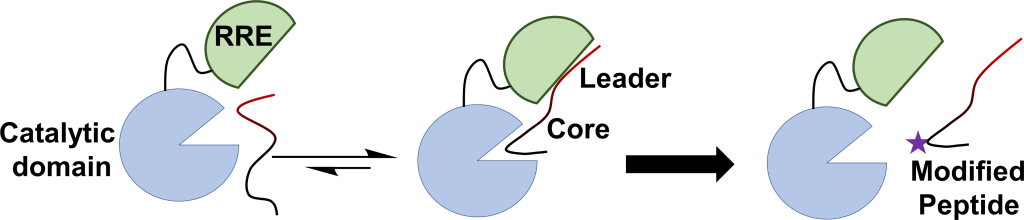
The Chekan Lab is focused on the discovery of new natural product biosynthetic pathways and uncovering the details of the enzymes that produce them. In particular, the Chekan Lab studies the Ribosomally Synthesized Post-translationally Modified Peptide (RiPP) class of natural products. As their name suggests, RiPPs take peptides made by the ribosome and then use tailoring enzymes to modify them into complex, bioactive structures. The final compounds contain a wide variety of bioactivities including antibacterial, antimalarial, metal chelating, and cell signaling. By combining bioinformatics, genome sequencing, protein biochemistry, structure elucidation, and chemical synthesis, the Chekan Lab aims to discover new RiPP compounds and connect known compounds back to their biosynthetic gene clusters.
In addition to discovery, the Chekan Lab is also interested in engineering new RiPP pathways, enzymes, and novel compounds with the goal of identifying new drug leads.