Publications
2025
Nguyen, A., Bunch, Z. L., Martinez-Aldino, I. Y., Rangel-Grimaldo, M., Tak, U., Thorstenson, J. C., Severn, M. M., Nakatsuji, T., Gallo, R. L., Graf, T. N., Oberlies, N. H., Chekan, J. R., Cech, N. B., and Horswill, A. R. (2025) An antimicrobial daptide from human skin commensal Staphylococcus hominis protects against skin pathogens. Nat. Commun. 16, 11459 (doi: 10.1038/s41467-025-66259-w)
Butler, K. S.*, Rajput, A.*, and Chekan, J. R. (2025) ThiF-Like Enzyme Chemistry in Primary and Secondary Metabolism. ChemBioChem. (doi: 10.1002/cbic.202500460)

Dommaraju, S. R.*, Kandy, S. K.*, Ren, H., Luciano, D. P., Fujiki, S., Sarlah, D., Zhao, H., Chekan, J. R.#, and Mitchell, D. A.# (2025) A versatile enzymatic pathway for modification of peptide C-termini. ACS Cent. Sci. (doi: 10.1021/acscentsci.5c01243)
Lima, S. T.*, Pasquale, M. A.*, Noyon, M. R. O. K., Clark, E. A., Laws, C. R., Hematian, S.#, and Chekan, J. R.# (2025) Peptide recognition sequence guides catalytic side chain cross-linking of plant peptides by copper-dependent cyclases. J. Am. Chem. Soc. 147, 20284–20293 (doi: 10.1021/jacs.4c15470)

Rajput, A.*, Butler, K. S.*, Springer, D. A., and Chekan, J. R. (2025) Indolylamide macrocyclization by a Streptococcus pneumoniae ThiF-like enzyme family member. Org. Lett. 27, 5765–5770 (doi: 10.1021/acs.orglett.5c01561)

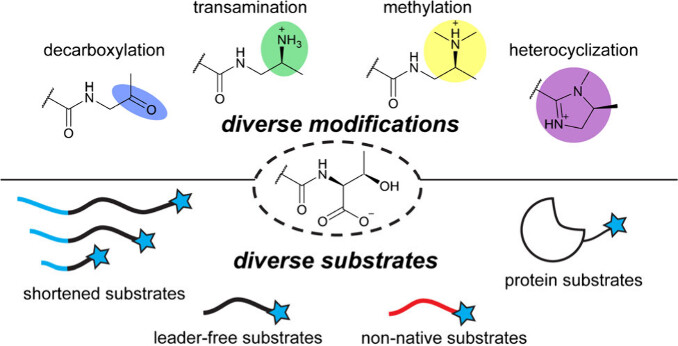
Smith, A. B., Ejindu, R. C., and Chekan, J. R. (2025) Engineering RiPP pathways: strategies for generating complex bioactive peptides. Trends Biochem. Sci. 50, 495–507 (doi: 10.1016/j.tibs.2025.04.001)

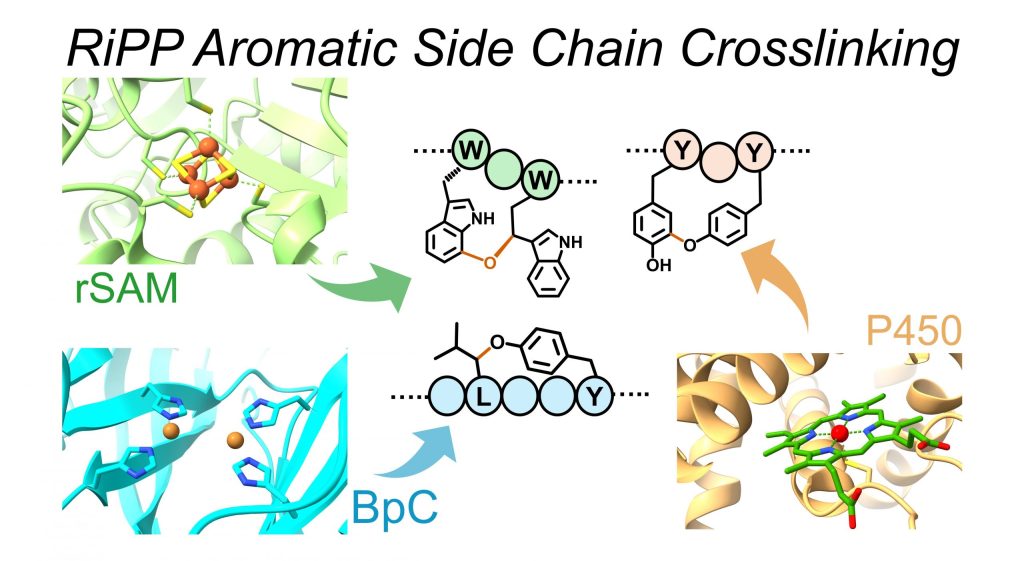
Kandy, S. K.*, Pasquale, M. A.*, and Chekan, J. R. (2025) Aromatic side-chain crosslinking in RiPP biosynthesis. Nat. Chem. Biol. (doi: 10.1038/s41589-024-01795-y)
2024
Hubrich, F., Kandy, S. K., Chepkirui, C., Padhi, C., Mordhorst, S., Moosmann, P., Zhu, T., Gugger, M., Chekan, J. R.# and Piel, J.# (2024) Ribosomal peptides with polycyclic isoprenoid moieties.Chem. (doi: 10.1016/j.chempr.2024.07.026)

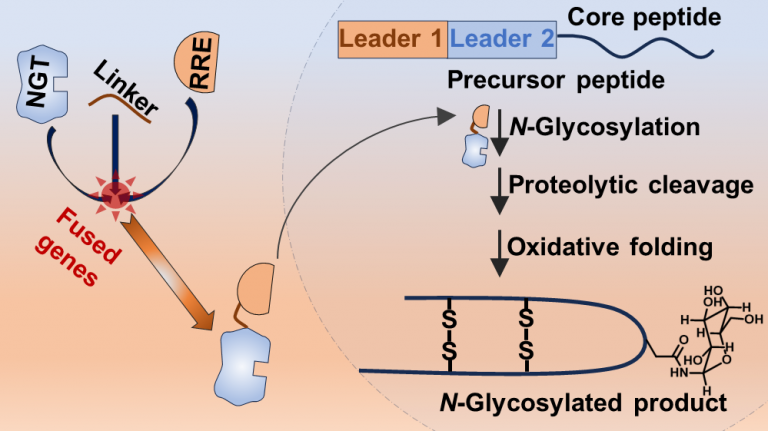
Smith, A. B., and Chekan, J. R. (2024) Targeted peptide modification using an engineered bacterial N-glycosyltransferase. ACS Catal. (doi: 10.1021/acscatal.4c01958)
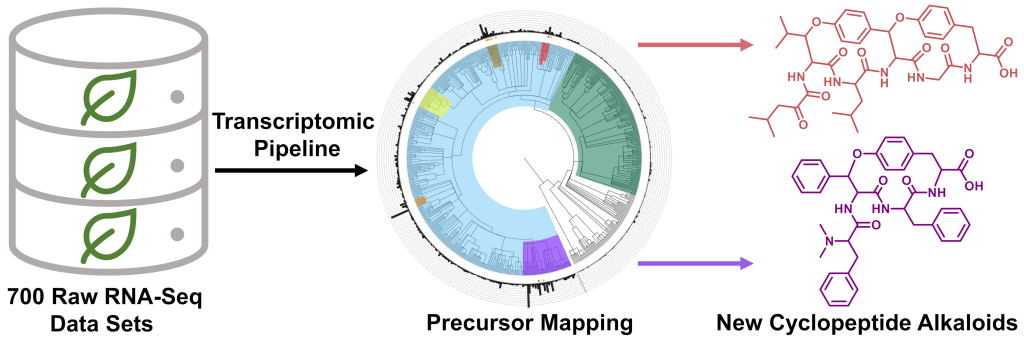
Kriger, D.*, Pasquale, M. A.*, Ampolini, B. G.*, and Chekan, J. R. (2024) Mining raw plant transcriptomic data for new cyclopeptide alkaloids. Beilstein J. Org. Chem. 20, 1548–1559 (doi: 10.3762/bjoc.20.138)
Pierre, H. C., Amrine, C. S. M., Doyle, M. G., Salvi, A., Raja, H. A., Chekan, J. R., Huntsman, A. C., Fuchs, J. R., Liu, K., Burdette, J. E., Pearce, C. J., and Oberlies, N. H. (2024) Verticillins: fungal epipolythiodioxopiperazine alkaloids with chemotherapeutic potential. Nat. Prod. Rep. (doi: 10.1039/d3np00068k)
Chekan, J. R.#, Mydy, L. S., Pasquale, M. A., and Kersten, R. D.# (2024) Plant peptides – redefining an area of ribosomally synthesized and post-translationally modified peptides. Nat. Prod. Rep. (doi: 10.1039/D3NP00042G)

2023
Cordoza, J. L., Chen, P. Y.-T., Blaustein, L. R., Lima, S. T., Fiore, M. F., Chekan, J. R., Moore, B. S., and McKinnie, S. M. K. (2023) Mechanistic and Structural Insights into a Divergent PLP-Dependent L-Enduracididine Cyclase from a Toxic Cyanobacterium. ACS Catal. 13, 9817–9828 (doi: 10.1021/acscatal.3c01294)
Gallardo, I. A., Todd, D. A., Lima, S. T., Chekan, J. R., Chiu, N. H., and Taylor, E. W. (2023) SARS-CoV-2 Main Protease Targets Host Selenoproteins and Glutathione Biosynthesis for Knockdown via Proteolysis, Potentially Disrupting the Thioredoxin and Glutaredoxin Redox Cycles. Antioxidants. 12, 559 (doi: 10.3390/antiox12030559)
Lima, S. T.*, Ampolini, B. G.*, Underwood, E. B.*, Graf, T. N., Earp, C. E., Khedi, I. C., Pasquale, M. A., and Chekan, J. R. (2023) A Widely Distributed Biosynthetic Cassette Is Responsible for Diverse Plant Side Chain Cross-Linked Cyclopeptides. Angew. Chem. Int. Ed Engl. (doi: 10.1002/anie.202218082)

2022
Smith, A. B., and Chekan, J. R. (2022) Engineering yeast for industrial-level production of the antimalarial drug artemisinin (Spotlight). Trends Biotechnol. (doi: 10.1016/j.tibtech.2022.12.007)
Kandy, S. K., and Chekan, J. R. (2022) Biosynthesis of the Plant-Produced Toxin Strychnine Elucidated (Highlight). Angew. Chem. Int. Ed Engl. 61, e202212301 (doi: 10.1002/anie.202212301)
Chen, P. Y.-T., Adak, S.,Chekan, J. R., Liscombe, D. K., Miyanaga, A., Bernhardt, P., Diethelm, S., Fielding, E. N., George, J. H., Miles, Z. D., Murray, L. A. M., Steele, T. S., Winter, J. M., Noel, J. P., and Moore, B. S. (2022) Structural Basis of Stereospecific Vanadium-Dependent Haloperoxidase Family Enzymes in Napyradiomycin Biosynthesis. Biochemistry. (doi: 10.1021/acs.biochem.2c00338)
Lima, S. T., Fallon, T. R., Cordoza, J. L., Chekan, J. R., Delbaje, E., Hopiavuori, A. R., Alvarenga, D. O., Wood, S. M., Luhavaya, H., Baumgartner, J. T., Dörr, F. A., Etchegaray, A., Pinto, E., McKinnie, S. M. K., Fiore, M. F., and Moore, B. S. (2022) Biosynthesis of Guanitoxin Enables Global Environmental Detection in Freshwater Cyanobacteria. J. Am. Chem. Soc. 144, 9372–9379 (doi: 10.1021/jacs.2c01424)
Steele, T. S., Brunson, J. K., Maeno, Y., Terada, R., Allen, A. E., Yotsu-Yamashita, M., Chekan, J. R.#, and Moore, B. S.# (2022) Domoic acid biosynthesis in the red alga Chondria armata suggests a complex evolutionary history for toxin production. Proc. Natl. Acad. Sci. 119, e2117407119 (doi: 10.1073/pnas.2117407119)
2021
Bauman, K. D., Butler, K. S., Moore, B. S.#, and Chekan, J. R.# (2021) Genome mining methods to discover bioactive natural products. Nat. Prod. Rep. (doi: 10.1039/d1np00032b)

Griffin, S. L., Chekan, J. R., Lira, J. M., Robinson, A. E., Yerkes, C. N., Siehl, D. L., Wright, T. R., Nair, S. K., and Cicchillo, R. M. (2021) Characterization of a Glyphosate-Tolerant Enzyme from Streptomyces svecius: A Distinct Class of 5-Enolpyruvylshikimate-3-phosphate Synthases. J. Agric. Food Chem. 69, 5096–5104 (doi: 10.1021/acs.jafc.1c00439)
Prior To Uncg
Chekan, J. R., Fallon, T. R., and Moore, B. S. (2020) Biosynthesis of marine toxins. Curr. Opin. Chem. Biol. 59, 119–129 (doi:10.1016/j.cbpa.2020.06.009)
Chekan, J. R., McKinnie, S. M. K., Noel, J. P., and Moore, B. S. (2020) Algal neurotoxin biosynthesis repurposes the terpene cyclase structural fold into an N -prenyltransferase. Proc. Natl. Acad. Sci. (doi:10.1073/pnas.2001325117)
Fiore, M. F., de Lima, S. T., Carmichael, W. W., McKinnie, S. M. K., Chekan, J. R., and Moore, B. S. (2020) Guanitoxin, re-naming a cyanobacterial organophosphate toxin. Harmful Algae. 92, 101737 (doi:10.1016/j.hal.2019.101737)
Chekan, J. R., Ongpipattanakul, C., and Nair, S. K. (2019) Steric complementarity directs sequence promiscuous leader binding in RiPP biosynthesis. Proc. Natl. Acad. Sci. U. S. A. 116, 24049–24055 (doi:10.1073/pnas.1908364116)
Chekan, J. R., Lee, G. Y., El Gamal, A., Purdy, T. N., Houk, K. N., and Moore, B. S. (2019) Bacterial tetrabromopyrrole debrominase shares a reductive dehalogenation strategy with human thyroid deiodinase. Biochemistry. 58, 5329–5338 (doi:10.1021/acs.biochem.9b00318)
Chekan, J. R.*, Ongpipattanakul, C.*, Wright, T. R., Zhang, B., Bollinger, J. M., Rajakovich, L. J., Krebs, C., Cicchillo, R. M., and Nair, S. K. (2019) Molecular basis for enantioselective herbicide degradation imparted by aryloxyalkanoate dioxygenases in transgenic plants. Proc. Natl. Acad. Sci. 116, 13299–13304 (doi:10.1073/pnas.1900711116)
Chekan, J. R., McKinnie, S. M. K., Moore, M. L., Poplawski, S. G., Michael, T. P., and Moore, B. S. (2019) Scalable biosynthesis of the seaweed neurochemical, kainic acid. Angew. Chemie Int. Ed. 58, 8454–8457 (doi:10.1002/anie.201902910)
Luhavaya, H., Sigrist, R., Chekan, J. R., McKinnie, S. M. K., and Moore, B. S. (2019) Biosynthesis of L-4-chlorokynurenine, an antidepressant prodrug and a non-proteinogenic amino acid found in lipopeptide antibiotics. Angew. Chemie Int. Ed. 58, 8394–8399 (doi:10.1002/anie.201901571)
Chekan, J. R., and Moore, B. S. (2018) Preparation and characterization of tetrabromopyrrole debrominase from marine proteobacteria. Methods Enzymol. 605, 253–65 (doi:10.1016/bs.mie.2018.01.031)
Brunson, J. K.*, McKinnie, S. M. K.*, Chekan, J. R., McCrow, J. P., Miles, Z. D., Bertrand, E. M., Bielinski, V. A., Luhavaya, H., Oborník, M., Smith, G. J., Hutchins, D. A., Allen, A. E., and Moore, B. S. (2018) Biosynthesis of the neurotoxin domoic acid in a bloom-forming diatom. Science. 361, 1356–8 (doi:10.1126/science.aau0382)
Chekan, J. R., and Moore, B. S. (2018) Biosynthesis of the antibiotic bicyclomycin in soil and pathogenic bacteria (Viewpoint). Biochemistry. 57, 897–898 (doi:10.1021/acs.biochem.7b01204)
Repka, L. M.*, Chekan, J. R.*, Nair, S. K., and van der Donk, W. A. (2017) Mechanistic Understanding of Lanthipeptide Biosynthetic Enzymes. Chem. Rev. 117, 5457–5520 (doi:10.1021/acs.chemrev.6b00591)
Olivares, P.*, Ulrich, E. C.*, Chekan, J. R.*, van der Donk, W. A., and Nair, S. K. (2017) Characterization of Two Late-Stage Enzymes Involved in Fosfomycin Biosynthesis in Pseudomonads. ACS Chem. Biol. 12, 456–463 (doi:10.1021/acschembio.6b00939)
Chekan, J. R., Koos, J. D., Zong, C., Maksimov, M. O., Link, A. J., and Nair, S. K. (2016) Structure of the lasso peptide isopeptidase identifies a topology for processing threaded substrates. J. Am. Chem. Soc. 138, 16452–16458 (doi:10.1021/jacs.6b10389)
Chekan, J. R.*, Cogan, D. P.*, and Nair, S. K. (2016) Molecular basis for resistance against phosphonate antibiotics and herbicides. Med. Chem. Commun. 7, 28–36 (doi:10.1039/C5MD00351B)
Peck, S. C., Chekan, J. R., Ulrich, E. C., Nair, S. K., and van der Donk, W. A. (2015) A common late-stage intermediate in catalysis by 2-hydroxyethyl-phosphonate dioxygenase and methylphosphonate synthase. J. Am. Chem. Soc. 137, 3217–20 (doi:10.1021/jacs.5b00282)
Chekan, J. R.*, Kwon, I. H.*, Agarwal, V.*, Dodd, D., Revindran, V., Mackie, R. I., Cann, I., and Nair, S. K. (2014) Structural and biochemical basis for mannan utilization by Caldanaerobius polysaccharolyticus strain ATCC BAA-17. J. Biol. Chem. 289, 34965–77 (doi:10.1074/jbc.M114.579904)
Zhang, M.*, Chekan, J. R.*, Dodd, D.*, Hong, P.-Y., Radlinski, L., Revindran, V., Nair, S. K., Mackie, R. I., and Cann, I. (2014) Xylan utilization in human gut commensal bacteria is orchestrated by unique modular organization of polysaccharide-degrading enzymes. Proc. Natl. Acad. Sci. U. S. A. 111, E3708–17 (doi:10.1073/pnas.1406156111)
Dunbar, K. L.*, Chekan, J. R.*, Cox, C. L., Burkhart, B. J., Nair, S. K., and Mitchell, D. A. (2014) Discovery of a new ATP-binding motif involved in peptidic azoline biosynthesis. Nat. Chem. Biol. 10, 823–9 (doi:10.1038/nchembio.1608)
Hung, J. E., Fogle, E. J., Garg, N., Chekan, J. R., Nair, S. K., and van der Donk, W. A. (2014) Chemical rescue and inhibition studies to determine the role of Arg301 in phosphite dehydrogenase. PLoS One. 9, e87134 (doi:10.1371/journal.pone.0087134)
Agarwal, V., Peck, S. C., Chen, J.-H., Borisova, S. A., Chekan, J. R., van der Donk, W. A., and Nair, S. K. (2014) Structure and function of phosphonoacetaldehyde dehydrogenase: the missing link in phosphonoacetate formation. Chem. Biol. 21, 125–35 (doi:10.1016/j.chembiol.2013.11.006)

